Before describing the PMF calculation in condensed phase systems, it can be useful to indicate the particular case of free energy calculations in small molecular systems. In the canonical ensemble (NVT) the Helmholtz free energy
Free energy calculations for small molecules:
Before describing the PMF calculation in condensed phase systems, it can be useful to indicate the particular case of free energy
calculations in small molecular systems.
In the canonical ensemble (NVT) the Helmholtz free energy ![]() is computed from the canonical partition function
is computed from the canonical partition function ![]()
| (2.114) |
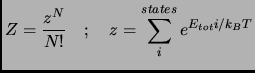 |
(2.115) |
In non-rigid condensed phase systems, the independent particle assumption ![]() is not valid, and the translational
partition function
is not valid, and the translational
partition function ![]() cannot be modeled as a free particle. Therefore, we will have to compute
the whole partition function through the phase space integral which in the NVT ensemble takes into account all the degrees
of freedom of the configuration space.
cannot be modeled as a free particle. Therefore, we will have to compute
the whole partition function through the phase space integral which in the NVT ensemble takes into account all the degrees
of freedom of the configuration space.
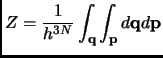 e e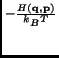 |
(2.117) |
The usual Hamiltonian expressions permits to separate the atomic momenta from the potential energy.
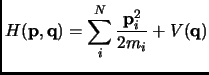 |
(2.118) |
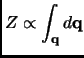 e e |
(2.119) |
We could consider our condensed phase system (solution or biomolecule) as a supermolecule and apply the same formula
than equations 1.116. However, when computing the vibrational motion of this supermolecule
the local and harmonic approximation is not valid since there are many minima separated by barriers lower than the k![]() T factor.
In consequence we still need to compute
the internal nuclear motion by MD or MC techniques until convergence of the integral of configuration.
T factor.
In consequence we still need to compute
the internal nuclear motion by MD or MC techniques until convergence of the integral of configuration.
After the assumption of Born-Oppenheimer approximation the nuclei have a classical behavior and it can be entirely reproduced by classical statistical mechanics. However, it has been seen that the absence of the zero point energy and the effects of quantized vibrational motion not contemplated in classical statistical mechanics can be a source of error, mainly when computing activation free energies in reactions involving hydrogen transfer [194].