



Next: Modulation of substrate activity
Up: Mandelate Racemase enzyme
Previous: Presentation of Mandelate Racemase
Contents
Introduction: Previous theoretical studies
Mandelate Racemase has already been studied in our group [212]
Before that, only a previous study by Alagona, Ghio and Kollman [232]
covered the Mandelate Racemase reactivity.
Other studies such as that from Maurice and Bearne [223]
perform geometry optimizations and electrostatic map potentials of several substrates
to elucidate which of the interacting groups of the substrate play an important role
in the active site. But they give no insight into the chemistry of the enzymatic reaction.
Kollman and co-workers
The work of Kollman et al. [232] is a very preliminary gas phase study where a small model of
active site is computed at HF/(STO-3G,3-21G and 6-31G) level.
This in vacuo study of the enzymatic reaction has some drawbacks such as the exclusion
of important residues in the model system. This is probably the reason why they observe no isomerization reaction.
They had important difficulties to obtain an optimized geometry that could reproduce
the X-ray active site, so they had to perform restricted geometry optimizations
in order to avoid artificial geometries and interactions. This fact gave also a too rigid
system unable to follow all the charge redistribution along the reaction path.
In section 2.4 we will perform similar gas-phase calculations and we will observe that
when the active site is represented with few atoms the model is not representative and it is unable to reproduce
the racemization reaction.
Garcia-Viloca et al.
We will briefly comment on the work carried out by Mireia Garcia-Viloca et al. some time ago [212].
This paper was done just before the author of this thesis started his work, so this can be taken as
a starting point of the whole thesis.
This is a QM/MM study of the racemization of vinylglycolate by Mandelate Racemase.
In table 2.1 vinylglycolate has already been shown as a possible substrate.
The reactivity is elucidated finding the possible intermediates by minimization of the energy function.
A coordinate scan between the intermediates is performed to have an approximation to the energy barrier of the reaction step.
We do not want to summarize all the study, but some conclusions stated there will be used in our results section 2.3.
This is the case for the reaction mechanisms encountered for vinylglycolate racemization.
It will be also important to know how the semiempirical Hamiltonian and the number of QM atoms were chosen.
Brief description of the reaction mechanisms
Three possible mechanisms are found.
Mechanism I and mechanism II involve six steps through six transition states and five intermediates each.
Both mechanisms require a proton transfer (the first step) from Lys164 or Glu317, respectively,
to a carboxylate oxygen of vinylglycolate through the corresponding hydrogen bond.
That proton transfer precedes the abstraction of the 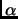 -proton (the second step),
so avoiding the accumulation of more than one negative net charge in the substrate.
-proton (the second step),
so avoiding the accumulation of more than one negative net charge in the substrate.
Figure 2.4:
A very simplified representation of the three mechanisms encountered in vinylglycolate racemization.
|
|
In contrast, a two-step mechanism (mechanism III) that takes place through a dianionic intermediate is also possible.
In this case, no previous proton transfer is required,
the conjugate acid of His297 now plays a stabilizing role together with the hydrogen bonds from both Lys164 and Glu317.
The three parallel mechanisms are competitive at room temperature, mec I being slower than the other two.
Choice of the QM Hamiltonian
Small models of the reaction were designed to select the semiempirical Hamiltonian.
They had compared the AM1 and PM3 performance with DFT results.
The energy profile for abstraction of the 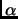 -proton of vinylglycolate by ethylamine
that models the residue Lys166 in the active site has been calculated at the different levels.
The comparison of geometries and energies gives the PM3 as the best semiempirical choice.
-proton of vinylglycolate by ethylamine
that models the residue Lys166 in the active site has been calculated at the different levels.
The comparison of geometries and energies gives the PM3 as the best semiempirical choice.
The X-ray crystal structure of the enzyme3.5 indicates that the
distance between the hexa-coordinated Mg ion and the oxygen atoms of the ligands is within the
range 2.0-2.5 Å (see figure 2.3 in page ![[*]](./icons/crossref.png) ).
However, the preliminary results using PM3 gave shorter distances with deviations
of 0.2-0.6 Å from the experimental. This is probably because PM3 parameters for magnesium have
been obtained mainly from magnesium halides and other small inorganic compounds data.
Then, Hutter et al.[234] developed new AM1 parameters for magnesium including
a wide variety of biologically relevant molecules that contain magnesium atoms with different coordinations.
The usage of AM1-SRP3.6 parameters for Mg and the rest of atoms in PM3 improves
significantly the results (the calculated distances are in the experimental range).
These same parameters have been applied successfully to other magnesium dependent enzymatic systems[189,158].
).
However, the preliminary results using PM3 gave shorter distances with deviations
of 0.2-0.6 Å from the experimental. This is probably because PM3 parameters for magnesium have
been obtained mainly from magnesium halides and other small inorganic compounds data.
Then, Hutter et al.[234] developed new AM1 parameters for magnesium including
a wide variety of biologically relevant molecules that contain magnesium atoms with different coordinations.
The usage of AM1-SRP3.6 parameters for Mg and the rest of atoms in PM3 improves
significantly the results (the calculated distances are in the experimental range).
These same parameters have been applied successfully to other magnesium dependent enzymatic systems[189,158].
Selection of the QM model
The final QM subsystem had been chosen after testing several sizes.
They looked for the model that minimized the polarization of the frontier link atoms.
In the end it included the substrate, the general acid based catalyst Lys166 and His297,
the charge stabilization residues Lys164, Glu317 and the magnesium cation with its additional four ligands.
The picture of the final QM subsystem is plotted in figure 2.3 in page ![[*]](./icons/crossref.png) .
.
Table 2.2:
Number of heavy atoms represented quantum
mechanically in each residue of the active center of the complex
MR-substrate
| Lys164 |
3 |
His297 |
5 |
| Lys166 |
4 |
Glu317 |
4 |
| Asp195 |
4 |
Substrate |
7 |
| Glu221 |
4 |
Mg |
1 |
| Glu247 |
5 |
H O O |
1 |
|
Besides, some residues have QM lateral chains larger than the ones usually adopted in simulations (see table 2.2).
That is, glutamic and aspartic are usually represented by ethanoic and lysine by ethylamine.
But the authors found that some residues could reproduce better the ab initio results by enlarging the QM lateral chain.
This is the case of Lys166 that is represented by propilamine, and Glu247 by propionate.




Next: Modulation of substrate activity
Up: Mandelate Racemase enzyme
Previous: Presentation of Mandelate Racemase
Contents
Xavier Prat Resina
2004-09-09
![\includegraphics[width=0.99\textwidth]{Figures/mechanism_vin.eps}](img455.png)