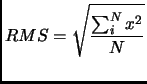- ... research.2.1
- Lots of names can be used to define the theoretical chemist research, e.g.
biophysics, molecular modeling, nano-materials, drug design, solid state physics, computational structural biology ...etc.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... core-electrons2.2
- Sometimes a screening or repulsive function is used
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... PM52.3
- The parameters of PM5
are not published and therefore unknown, these kind of commercial initiatives does not help very much
to the progress in computational chemistry
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... backward.2.4
- It can be shown that while expression 1.67 is a first order approximation in a Taylor expansion,
equation 1.68 is the second order one.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... point)2.5
- Since the numerical value of the norm depends on the dimension of the vector, the root mean square (RMS)
is a more adequate magnitude
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...simons_jpc83,banerjee_jpc85.2.6
- In the original publication this method was designed for wavefunction
optimization rather than molecular geometries
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...fukui_acr81
2.7
- Some authors consider that the MEP is only when the path is
computed in mass-weighted coordinates, that is, MEP and IRC are synonyms
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... derivatives2.8
-
The conjugate peak refinement [146] represents the Hessian matrix in its equations by the identity matrix
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... atoms2.9
-
A computer of 512 MB of free memory can store 1024*1024*512=536 870 912 bytes
 67 108 864 double precision real numbers
which in triangular form (n(n+1)/2) can build a matrix of dimension 11584.
67 108 864 double precision real numbers
which in triangular form (n(n+1)/2) can build a matrix of dimension 11584.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... motion2.10
- Other formulations of classical mechanics alternative to the Newton's are also used [166,167]
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... eliminated2.11
- It has been seen that inclusion of the mean
electrostatic field without properly accounting for fluctuations in this field leads to distortions in the simulation [186]
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... unique2.12
- For example, in chapter 4
we will use an additional polynomic function
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... space2.13
- Note that consider a saddle point in
the configuration space as the transition state may not always be correct
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... enantiomers3.1
- The epimerases family are closely related since they interconvert the two
diastereoisomers
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...gerlt_bio95.3.2
-
Turnover number is the
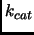 often applied to enzyme catalyzed reactions.
It represents the maximum number of substrate molecules (when the substrate concentration
is very high) which can be converted to products per molecule of enzyme per unit time
often applied to enzyme catalyzed reactions.
It represents the maximum number of substrate molecules (when the substrate concentration
is very high) which can be converted to products per molecule of enzyme per unit time
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... MR)3.3
- Enzyme Commission (EC) classifies MR in the main class 5 of isomerases
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
binding3.4
- This new experimental
result seems to be against the hypothesis that a formation of a strong and short hydrogen bond between mandelate
and Glu317 is essential for the catalysis effect[225]
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... enzyme3.5
- PDB code 1MNS[233]
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... AM1-SRP3.6
- Although the AM1 parameters optimized for magnesium can be used in a wide variety
of molecules, since they are not the standard ones we use the term SRP to indicate it.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...merzbanci_jpc963.7
- This strategy is not necessary when a force field has an integer charge for every
small group of atoms (e.g. CHARMM force field) or when the QM/MM frontier already contemplates the correction (GHO frontier, for example).
This method will not be employed in chapter 4
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... I3.8
-
It is clearly a dianionic since no proton transfer occurs from Lys164 or Glu317
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... site3.9
-
This is a very common situation. Two minima can differ in some units of kcal/mol without any significant geometry difference
in the active site.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... regions3.10
-
A normal mode analysis gives a high number of low frequencies.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... mechanism 3.11
- This problem
exists also in QM/MM when we have to treat some residues with MM potentials.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... charge4.1
- http://www.bath.ac.uk/ chsihw/grace/grace.html
A similar package [161] is implemented in the Carr-Parrinello package: http://www.cpmd.org
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... search4.2
-
In LAPACK library there are several algorithms that permit this more economic diagonalization[246].
We will use the term partial for those diagonalization methods that extract a limited number of eigenpairs but
require the whole matrix in memory. The label iterative will be used for the methods studied in section 3.4
where the memory storage of the matrix is avoided
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... core4.3
- the
nomenclature core/environment used here will appear in the micro-iterations scheme.
But in this case it only indicates the partition in a QM and MM region. So, the whole set of atoms are moved in a unique process
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... zone4.4
-
This is an important difference with the IMOMM-ONIOM scheme [108,162] where no explicit
polarization of the MM part is included; and probably this is why a full and yet cheap minimization was done at every core step
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... work.4.5
-
This relative uncertainty will be exemplified in the appendix section testing different compilers and
several optimization levels in compilation
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... iteration4.6
-
The partial diagonalization process extract the 5 lowest eigenpairs to calculate the displacement in RFO
and to check the curvature in the current region of the PES.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... diagonalization.4.7
-
Obviously, since the Hessian is a symmetric matrix we are always working with the triangular part
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... problem4.8
- See truncated Newton Rapshon as another alternative [156,157]
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... function4.9
- This technique is in the inverse direction of the Hartree-Fock-Roothan equations where
a minimization of the energy leads to a diagonalization of the Fock matrix
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... encountered5.1
- See the appendix for the source code developed in order to obtain the PMF profile
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ...
times5.2
- Personal communication from Prof. Jiali Gao
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
- ... GNUA.1
-
GNU: http://www.gnu.org/
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.